-
About
- Kyoto Prize
-
Research Grants
-
Social Contributions
- Events
- News
This website uses cookies to improve the user experience. If you continue on this website, you will provide your consent to our use of cookies.
About
Research Grants
Social Contributions

InaRIS Fellow (2021-2030)
Professor,Research Center for Advanced Science and Technology, The University of Tokyo*Profile is at the time of the award.
2021InaRISBiology & Life sciences
Last year I opened my own laboratory, and this generous grant will allow me to devote my time to research. I am very grateful for support from the Inamori Research Institute for Science (InaRIS) for the next 10 years. The timing is extremely fortunate as I am at a turning point in my research life. I would like to engage in curiosity-driven research about the action mechanisms of proteins and nucleic acids as my research lifework.
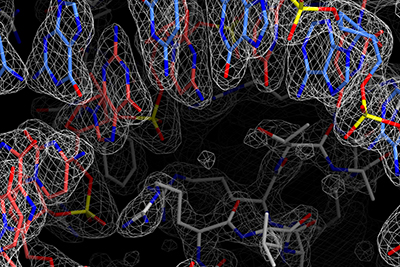
We determined the cryo-EM structures of several novel Cas enzymes, including Cas12c2, Cas12f, and Cas13bt3, elucidating their diverse mechanisms of action. In 2020, I established my own lab at The University of Tokyo and elucidated the cryo-EM structures of the new Cas enzyme Cas7-11-Csx29 and IscB, the ancestor of Cas9. In addition, I initiated research on DNA recombinases, determining the cryo-EM structures of the serine recombinase Bxb1 and a unique RNA-guided IS110 recombinase. These studies provide mechanistic insights into DNA recombination and establish a foundation for advancing genome engineering technologies.
In the CRISPR-Cas adaptive immune system, diverse Cas enzymes are responsible for defense against foreign nucleic acids and form ribonucleoprotein complexes with their cognate guide RNA to recognize and cleave target nucleic acids (DNA or RNA). Using cryo-electron microscopy, we have determined the structures of several novel Cas enzymes, including Cas12c2 (Kurihara et al., Mol Cell 2022), Cas12f (Takeda et al., Mol Cell 2021), and Cas13bt3 (Nakagawa et al., Mol Cell 2022), elucidating their distinct mechanisms of action. In 2020, I established a laboratory at The University of Tokyo and have been promoting structural and functional studies of the novel Cas enzyme complex Cas7-11-Csx29 and IscB, the likely ancestor of Cas9 (Kato et al., Cell 2022; Kato et al., Science 2022; Kato et al., Nat Commun 2022). We also initiated research on DNA recombinase such as the serine recombinase Bxb1 and the novel bridge RNA-guided IS110 family recombinase.
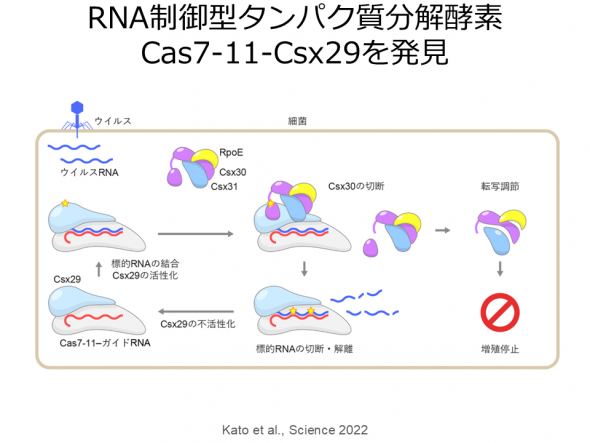
In the type III-E CRISPR-Cas system, Cas7-11 functions as an RNA-guided RNA nuclease that specifically cleaves single-stranded RNA targets at two sites. While Cas7-11 could be used as a highly specific RNA-targeting tool with low cytotoxicity, its RNA cleavage mechanism was enigmatic. Furthermore, the III-E type CRISPR-Cas locus encodes uncharacterized proteins, such as Csx29, Csx30, Csx31, and RpoE, suggesting that these proteins collaborate in antiviral defense. We determined the cryo-EM structure of the Cas7-11-guide RNA-target RNA complex and elucidated the Cas7-11-mediated RNA cleavage mechanism (Kato et al., Cell 2022). The structure revealed that Cas7-11 comprises four Cas7 domains (Cas7.1-Cas7.4), a Cas11 domain, an INS domain, a CTE domain, and four linker regions. Our biochemical experiments showed that Cas7-11 cleaves the phosphodiester bonds between bases 3-4 and 9-10 of the target RNA. Based on the structural information, we engineered a compact Cas7-11 variant (Cas7-11S), which can be efficiently delivered to target tissues as a knockdown tool (Kato et al., Cell 2022).
Furthermore, our biochemical analyses revealed that when the target RNA binds to the Cas7-11-guide RNA-Csx29 complex, Csx30 is cleaved by Csx29 into two fragments (Kato et al., Science 2022). The cryo-EM structures of the Cas7-11-guide RNA-Csx29 complex and the Cas7-11-guide RNA-Csx29-target RNA complex revealed that the binding of target RNA to the complex induces a structural change in the Csx29 active site, allowing Csx30 recognition and cleavage. These findings demonstrated that the Cas7-11-Csx29 complex is an unprecedented RNA-guided nuclease-protease complex with both RNA and Csx30 cleavage activities. Furthermore, we found that overexpression of Csx30 in Escherichia coli inhibits bacterial growth and that Csx30, Csx31, and RpoE form a complex, suggesting that in the type III-E system, the Cas7-11-Csx29 complex cleaves viral RNA and endogenous Csx30, thereby inhibiting the growth of infected cells through RpoE-mediated transcriptional regulation (Kato et al., Science 2022).
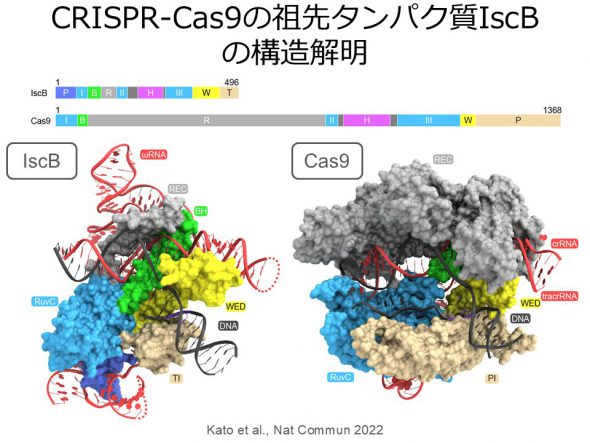
The RNA-guided DNA nuclease Cas9 is widely used in genome editing. Recent studies have indicated that Cas9 evolved from the transposon-encoded RNA-guided DNA nuclease IscB. However, the details of this molecular evolution remain unclear. To explore this, we determined the cryo-EM structure of the IscB-guide RNA-target DNA complex. A structural comparison of IscB and Cas9 suggested that the acquisition of protein domains and the miniaturization of the guide RNA contributed to the molecular evolution of CRISPR-Cas9 (Kato et al., Nat Commun 2022).
Recombinases are the most abundant proteins in nature and are classified into transposases and integrases, performing diverse functions across species from viruses to higher eukaryotes. The phage-derived large serine recombinase Bxb1 has been utilized in genome engineering technologies, as it catalyzes recombination between two DNA molecules with specific sequences (attP and attB). Serine recombinases have been studied since the 1980s, and a DNA recombination mechanism involving rotation has been proposed. However, their detailed mechanisms remain unclear. To address this, we determined the cryo-EM structure of the Bxb1-DNA complex, providing visual evidence for its unique DNA recombination mechanism.
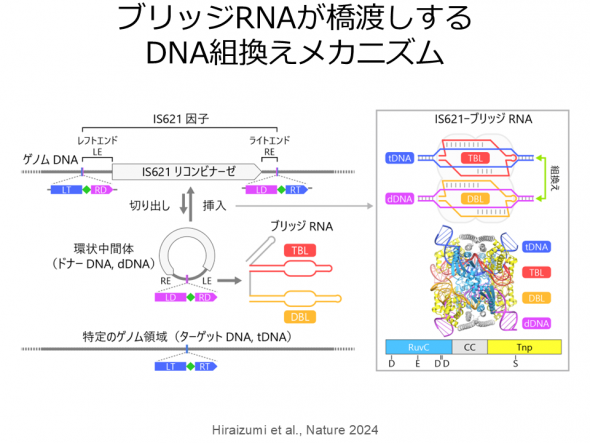
We also investigated the structure and function of a novel RNA-guided IS621 recombinase, which belongs to the IS110 family of recombinases. Our biochemical and cellular analyses revealed that the IS621 recombinase forms a complex with a bridge RNA and catalyzes recombination between donor DNA and target DNA that have sequences complementary to the guide segments in the bridge RNA. Furthermore, we determined the cryo-EM structure of the IS621 recombinase-bridge RNA-donor DNA-target DNA complex, elucidating the molecular mechanism of unprecedented RNA-guided DNA recombination. IS110 family recombinases are expected to be used as new genome engineering tools, and this structural information will serve as a foundation for the molecular engineering of IS110 recombinases.
CRISPR-Cas is a “robust” system responsible for acquired immune system in bacteria. When a bacterium is exposed to a viral infection, part of the viral sequence is incorporated into the CRISPR region of the bacterial genome, and when the same virus re-invades, the Cas enzyme uses the “memory” of viral sequence stored on CRISPR to cleave the invader’s genome. One of the Cas enzymes, Cas9, is an RNA-dependent DNA cleaving enzyme that uses an arbitrary RNA sequence as a template (guide RNA) to cleave the target DNA sequence at any desired point. Genome editing using CRISPR-Cas9 is a revolutionary technology that enables precise modification of genome sequences, the “blueprint of life”, and offers a wide range of applications from basic molecular biology research to drug discovery, gene therapy, and crop breeding. It is also fresh in our minds that it was the subject of last year’s Nobel Prize in Chemistry.
Dr. Nishimasu has made remarkable breakthroughs in understanding and applying the structure and function of the CRISPR-Cas system from the viewpoint of structural biology. For example, he was the first in the world to determine the crystal structure of the Streptococcus pyogenes Cas9-guide RNA-target DNA complex, which is widely used for genome editing. Furthermore, through the structure-guided protein engineering, he successfully redesigned the three-dimensional structure of Cas9 based on the structure he discovered and extended the range of application of genome editing technology fourfold. In addition, through collaboration with experts in single-molecule observation, he has succeeded in capturing a movie of the process of DNA cleavage by Cas9.
However, Cas enzymes are diverse, and there are many other types of Cas proteins besides Cas9. The function of many Cas enzymes is still not well understood. Recently, Dr. Nishimasu used cryo-electron microscopy to determine the structure of the guide RNA-target DNA complex of Cas12f, the smallest known Cas enzyme. Through structure-function analyses of Cas12f, he revealed a completely new mode of action of Cas enzyme.
Dr. Nishimasu’s research proposal aims to elucidate the operating principles of these Cas enzymes, whose functions are unknown, by utilizing his strengths in structural analysis of protein-RNA complexes and enzymological approaches. Furthermore, by teaming up with experts in marine metagenomics, he aims to search for completely new types of Cas enzymes from microorganisms that grow “flexibly” and “robustly” in extreme conditions such as the deep sea, and to elucidate their structures and functions. Microorganisms that adapt to and grow in extreme environments such as high temperature and high pressure are the frontier for the search for new enzymes: A great example includes Taq polymerase from a thermophilic bacterium has brought about the generalization of polymerase chain reaction (PCR) and innovation in research and diagnosis in medical biology.
Dr. Nishimasu is a rising star in structural biology who has just launched his own laboratory in the summer of 2020. Therefore, with the 10-year support of the InaRIS Fellowship, Dr. Nishimasu can pursue his bold and innovative ideas to advance the world of Cas enzymes . Furthermore, we hope that the fellowship will lead to the discovery of novel protein-RNA complex enzymes beyond Cas enzymes, and contribute to new developments in biology through the elucidation of their structures and operation mechanisms.
The Inamori Foundation announced the 2025 fellows for the Inamori Research Institute for Science (InaRIS) Fellowship Program on March 14, 2025.

The Inamori Foundation started to accept applications for the 2025 Inamori Research Institute for Science (InaRIS) Fellowship on May 20, 2024.

On April 20, the 2024 InaRIS Fellowship Award Ceremony was held at a hotel in Kyoto.

The Inamori Foundation announced the 2024 fellows for the Inamori Research Institute for Science (InaRIS) Fellowship Program on March 15, 2024. This year, we will welcome two new fellows, Hiroshi Suzuki (Professor, Graduate School of Medicine, Nagoya University) and Ayuko Hoshino (Professor, Research Center for Advanced Science and Technology, The University of Tokyo), who were...

The Inamori Foundation started to accept applications for the 2023 Inamori Research Institute for Science (InaRIS) Fellowship on May 23, 2022.

From the left: Executive Managing Director Shoichi Himono, InaRIS Chair Shigetada Nakanishi, the 2023 Fellows Yoshihiro Tanaka and Yasutaka Kamei, and Chair of Selection Committee Hiroto Yasuura On April 22, the InaRIS Fellowship Award Ceremony was held at a hotel in Kyoto. The 2023 InaRIS Fellows were recruited under the theme “Transcending the Future with...
The Inamori Foundation announced the 2023 fellows for the Inamori Research Institute for Science (InaRIS) Fellowship Program on March 17, 2023.

The Advisory Board Meeting of the Inamori Research Institute for Science (InaRIS) Fellowship Program was held at the Inamori Foundation in Kyoto, on October 4. There the committee members and the fellows had a discussion over their research themes. The InaRIS Fellowship Program was established in 2019 to encourage researchers to pursue their curiosity...

On April 23, InaRIS Fellowship Award Ceremony was held at a hotel in Kyoto for the first time since the establishment of the Fellowship in 2019.

The Inamori Foundation announced the 2022 fellows for the Inamori Research Institute for Science (InaRIS) Fellowship Program on March 18, 2022.

The Advisory Board Meeting of Inamori Research Institute for Science (InaRIS) Fellowship Program was held online on October 3 for the Committee members and the fellows to have a discussion over their research themes.

The Inamori Foundation started to accept applications for the 2022 Inamori Research Institute for Science (InaRIS) Fellowship on May 21, 2021.
On March 19, 2021, the Inamori Foundation announced the two 2021 fellows for the Inamori Research Institute for Science (InaRIS) Fellowship Program, Hiroshi Nishimasu (Professor, Research Center for Advanced Science and Technology, The University of Tokyo) and Yoshifumi Yamaguchi (Professor, Institute of Low Temperature Science, Hokkaido University) were selected from 66 applicants. InaRIS Fellowship Program...

Inamori Research Institute for Science (InaRIS) Fellowship Program is a research grant program that allows researchers to pursue challenging projects over a 10-year period. The Advisory Board Meeting of InaRIS Fellowship Program was held on October 26 at the Inamori Foundation in Kyoto to have a discussion over the fellows’ research themes. InaRIS Fellowship Program...

The Inamori Foundation started to accept applications for the 2021 Inamori Research Grants and Inamori Research Institute for Science (InaRIS) Fellowship on May 21. The Inamori Research Grants Program was established in 1985 to foster diverse and creative research activities by providing opportunities to challenge many potential themes. This program has little scrutiny on how...

On April 10, 2020, the Inamori Foundation announced the 2020 fellows for the Inamori Research Institute for Science (InaRIS) Fellowship Program. As the first two InaRIS fellows for this new program, Tadashi Takayanagi (Professor, Yukawa Institute for Theoretical Physics, Kyoto University) and Atsushi Noguchi (Associate Professor, Graduate School of Art and Sciences, The University of...

The InaRIS Fellowship Program grants 10 million yen per year over 10 years (totaling 100 million yen) to researchers engaged in investigations into topics with high potential based on a grand vision, providing them with opportunities to indulge themselves in research activities and pursue the possibility of triggering a quantum leap in science. The keyword...

The Inamori Foundation starts to accept registration for the Inamori Grants and Inamori Research Institute for Science (InaRIS) Fellowship.

Today, the Inamori Foundation announced an establishment of the new grants program: “Inamori Research Institute for Science (InaRIS) Fellowship Program” in the press conference at Ministry of Education, Culture, Sports, Science and Technology.
Biology & Life sciences